torchbox.ml package
Submodules
torchbox.ml.reduction_pca module
- torchbox.ml.reduction_pca.pca(x, sdim=-2, fdim=-1, npcs='all', eigbkd='svd')
Principal Component Analysis (pca) on raw data
- Parameters:
x (Tensor) – the input data
sdim (int, optional) – the dimension index of sample, by default -2
fdim (int, optional) – the dimension index of feature, by default -1
npcs (int or str, optional) – the number of components, by default
'all'eigbkd (str, optional) – the backend of eigen decomposition,
'svd'(default) or'eig'
- Returns:
U, S, K (if
npcsis integer)- Return type:
tensor
Examples

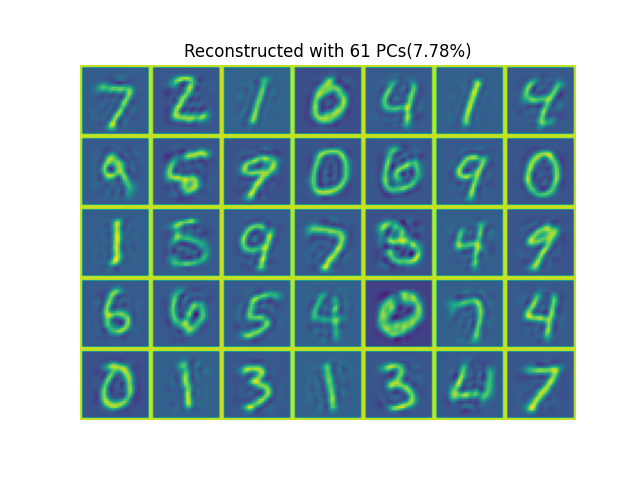
The results shown in the above figure can be obtained by the following codes.
rootdir, dataset = '/mnt/d/DataSets/oi/dgi/mnist/official/', 'test' x, _ = tb.read_mnist(rootdir=rootdir, dataset=dataset, fmt='ubyte') print(x.shape) N, M2, _ = x.shape x = x.to(th.float32) pcr = 0.9 u, s = tb.pca(x, sdim=0, fdim=(1, 2), eigbkd='svd') k = tb.pcapc(s, pcr=pcr) print(u.shape, s.shape, k) u = u[..., :k] y = x.reshape(N, -1) @ u # N-k z = y @ u.T.conj() # z[z<0] = 0 z = z.reshape(N, M2, M2) print(tb.nmse(x, z, dim=(1, 2))) xp = th.nn.functional.pad(x[:35], (1, 1, 1, 1, 0, 0), 'constant', 255) zp = th.nn.functional.pad(z[:35], (1, 1, 1, 1, 0, 0), 'constant', 255) plt = tb.imshow(tb.patch2tensor(xp, (5*(M2+2), 7*(M2+2)), dim=(1, 2)), titles=['Orignal']) plt = tb.imshow(tb.patch2tensor(zp, (5*(M2+2), 7*(M2+2)), dim=(1, 2)), titles=['Reconstructed with %d PCs(%.2f%%)' % (k, 100*k/u.shape[0])]) plt.show() u, s = tb.pca(x.reshape(N, -1), sdim=0, fdim=1, npcs=2, eigbkd='svd') print(u.shape, s.shape) y = x.reshape(N, -1) @ u # N-k z = y @ u.T.conj() z = z.reshape(N, M2, M2) print(tb.nmse(x, z, dim=(1, 2)))
- torchbox.ml.reduction_pca.pcapc(s, pcr=0.9)
get principal component according to the ratio of variance
- Parameters:
s (Tensor) – eigenvalues
pcr (float, optional) – the ratio of variance, by default 0.9
- torchbox.ml.reduction_pca.pcat(x, sdim=-2, fdim=-1, isnorm=True, eigbkd='svd')
gets Principal Component Analysis transformation
- Parameters:
x (Tensor) – the input data
sdim (int, optional) – the dimension index of sample, by default -2
fdim (int or tuple, optional) – the dimension index of feature, by default -1
isnorm (bool, optional) – whether to normalize covariance matric with the number of samples, by default True
eigbkd (str, optional) – the backend of eigen decomposition,
'svd'(default) or'eig'
- Returns:
the PCA transformation matrix, the eigenvalues
- Return type:
tensor